Home Research
Mentor/s:
José García Arrarás, Ph.D.
Project Title:
Gene Characterization of Holothurian Radial Nerve Cord
Project Description:
As a continuation of the single-nuclei RNA sequencing project done during the summer, I am currently part of a project concentrated on characterizing the genes present in the RNC during the normal and regenerating process using HCR-FISH. My target gene population is Sox9, which has been shown to have different marking capacities in model organisms. I’m also working on a bioinformatics project focused on validating and identifying different prominin sequences found in sequencings of Holothuria glaberrima.
Summer Research
Mentor/s:
José García Arrarás, Ph.D.
Project Title:
Single-nuclei RNA-seq of Holothurian Radial Nerve Cord
Project Description:
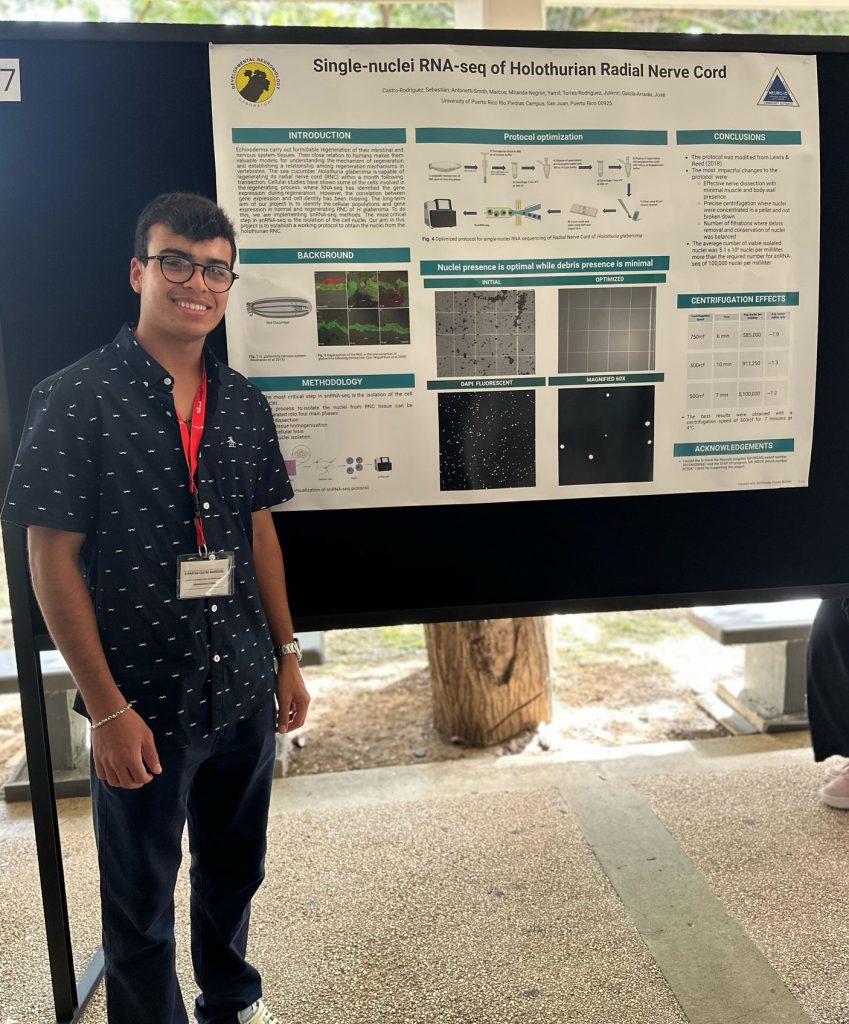
Echinoderms carry out formidable regeneration of their intestinal and nervous system tissues. Their close relation to humans makes them valuable models for understanding the mechanisms of regeneration and establishing a relationship between regeneration mechanisms in vertebrates. The sea cucumber Holothuria glaberrima is capable of regenerating its radial nerve cord (RNC) within a month following transection. Cellular studies have shown some of the cells involved in the regenerating process where RNA-seq has identified the gene expression during regeneration. However, the correlation between gene expression and cell identity has been missing. The long-term aim of our project is to identify the cellular populations and gene expression in normal and regenerating RNC of H. glaberrima. To do this, we are implementing single-nuclei RNA-sequencing (snRNA-seq) methods. The most critical step in snRNA-seq is the isolation of the cell nuclei. Our aim of establishing a working protocol to obtain the nuclei from the holothurian RNC has been achieved. The process to isolate the nuclei from RNC tissue can be separated into four main phases (1) dissection, (2) tissue homogenization, (3) cellular lysis, and (4) nuclei isolation. To optimize the protocol, we used different douncer homogenizers, managed the tissue at different temperatures, revised the number of filtrations, and changed the centrifugation parameters. The experiment was carried out successfully with an average number of 5,100,000 viable nuclei per milliliter, more than the required number for snRNA-seq of 100,000 nuclei. The data is in the process of being analyzed. To further the analysis, HCR-FISH is being used to validate cell populations found in the sequencing.
Summer Research
Mentor/s:
Paschalis Kratsios, Ph.D.
Project Title:
Molecular mechanisms of interneuron development in touch-evoked escape response
Project Description:
PENDING